Mechanisms of C3G-Rap1 signaling in physiology and diseases |
C3G is a guanine nucleotide exchange factor (GEF) that activates the small GTPase Rap1; doing so, C3G regulates cell
adhesion and other cellular functions. Before stimulation, resting C3G is in an inactive state with very low GEF activity.
We study how C3G is maintained in an inactive state and how it is activated.
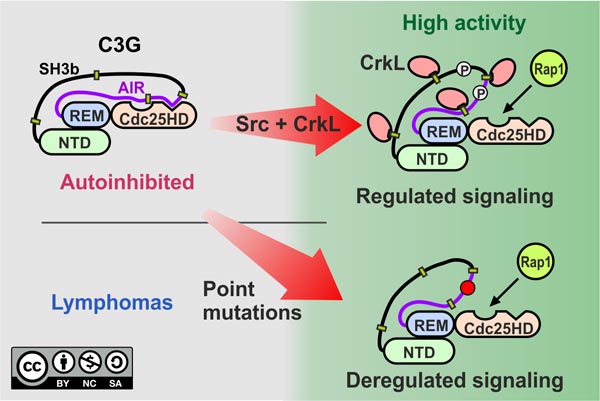
We have described in detail the mechanisms of autoregulation of C3G. The GEF activity of C3G resides in the Cdc25
homology domain (Cdc25HD). We have shown that C3G is regulated by two intramolecular interactions. A region of the central
SH3b domain binds to the Cd25HD and blocks the GEF activity; hence, we named that segment the autoinhibitory region or AIR.
Disruption of the AIR/Cdc25HD interaction is sufficient to fully activate C3G. This led us to identify two missense
mutations in the AIR that had been described in human lymphomas (Y554H and M555K) that cause constitutive activation of
C3G (Carabias et al, Sci Signal 2020). We propose that
C3G-activating mutations are a mechanism of cancer cells to acquire abnormal Rap1 signaling
(Carabias et al, Mol Cell Oncol. 2021).
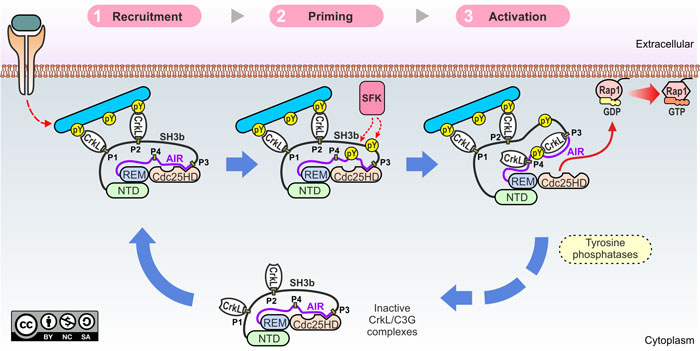
We have also elucidated the mechanisms of physiological C3G activation (Rodríguez-Blázquez, Carabias et al. Cell Commun Signal 2023). C3G is activated by stimuli that operate through tyrosine kinases, and this process requires the Crk adaptor proteins. C3G has four Pro-rich motifs (P1-P4) that are binding sites for the SH3N domain of CrkL and CrkII. We have shown that sites P1 and P2 are required for the translocation of CrkL-C3G complexes to the plasma membrane. However, activation requires binding of CrkL to mainly the P3 and secondarily P4 sites. We have also shown that tyrosine phosphorylation of C3G is necessary but not sufficient for activation.
How the guanine nucleotide exchange factor C3G is activated in cells from Research Square on Vimeo.
The multistep mechanisms of C3G activation is summarized in the following figure:
Funding:
Our work on C3G-Rap1 signaling was and is supported by the Spanish Ministry of Science and Innovation (MCIN), Agencia Estatal de Investigación (AEI), MCIN/ AEI/10.13039/501100011033/ and by European Regional Development Fund "ERDF a way of making Europe" (grants PID2019-105763GB-I00 and PID2022-136322NB-I00). This project has also received funding from Junta de Castilla y León and ERDF (grants SA017U16 and SA078P20).


Architecture and regulation of hemidesmosomes |
Hemidesmosomes are protein complexes that mediate the stable attachement of basal cells to the underlying basement
membrane in epithelial tissues, such as the skin. They provide a link between the extracellular matrix and the intermediate
filament cytoskeleton. Hemidesmosomes contain the following proteins: integrin α6β4, plectin,
BP230 (also known as BPAG1e), BP180 (known as Collagen XVII), and the tetraspanin CD151.
We study the structure of the proteins that form the hemidesmosomes and the structural basis of the interactions that
they establish. We also aim at understanding how hemidesmosomal proteins are altered in a type of blistering diseases
named epidermolysis bullosa.
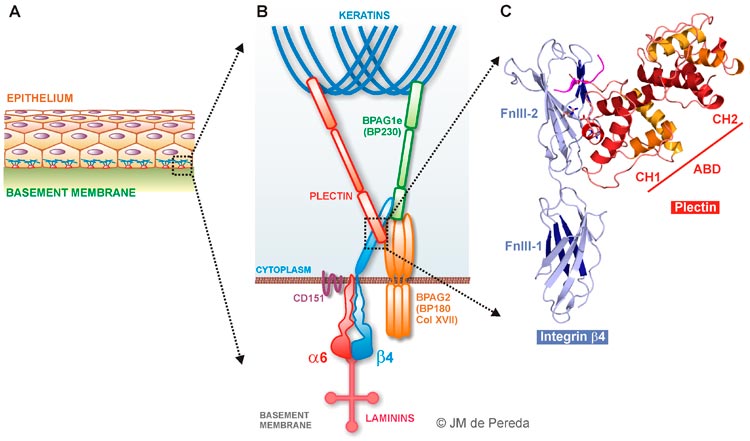
Schematic representation of the components of the hemidesmosomes and the interactions that they stablish with each other.
Integrin α6β4
Integrins are a family of hetero-dimeric transmembrane receptors that mediate cell adhesion and bi-directional
signal transduction. The α6β4 integrin is a laminin receptor.
The β4 subunit has an unusual cytoplasmic region (~1000 residues) that contains a Calx-β domain and
four "fibronectin type III" domains (FnIII-1 to FnIII-4).
We have determined the 3D structure of all these five globular domains.
These include the crystal structures of the Calx-β domain (
Alonso-García et al, Acta Cryst D 2009), the first pair of FnIII domain (FnIII-1,2)
(de Pereda et al, EMBO J 1999),
and the individual structures of the FnIII-3 and FnIII-4 domains (
Alonso-García et al, Acta Cryst D 2015).
To elucidated the structure of the FnIII-3,4 region we have used hybrid methods, in which we combined the crystal structures
of the FnIII-3 and FnIII-4 with SAXS, and double electron-electron resonance (DEER-EPR) data (
Alonso-García et al Acta Cryst D 2015).
Our contributions to the study of the structure of the integrin β4 subunit are summarized in the next figure:
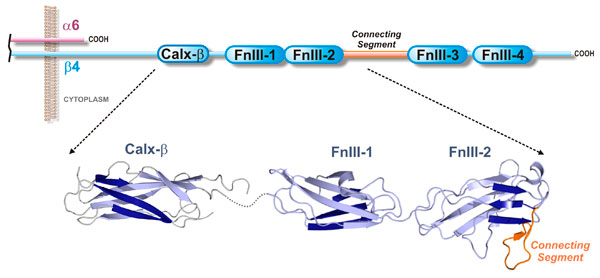
Structure of the cytoplasmic tails of integrin α6β4.
Plectin
Plectin belongs to the plakin protein family of cytoskeletal linkers. In hemidesmosomes plectin provides
a direct link between the integrin α6β4 and the cytokeratin filaments.
Plectin is a large protein (~500 kD) with a tripartite organization: it contains N- and C-terminal regions
that harbor protein-protein interaction sites and a central rod domain involved in oligomerization.
We have extensively characterized the N-terminal region of plectin. This segment consists of an actin
binding domain (ABD) and a region termed the plakin domain that is conserved in most plakins.
We have solved the crystal structure of the isolated ABD
(García-Álvarez et al, Structure 2003).
We have also solve the 3D structure of the ABD bound to the FnIII-1,2 of integrin α6β4.
Our structure of the plectin-α6β4 comlex revealed key determinants for the interaction.
It also showed that two single amino-acid substitutions in the integrin that cause epidermolysis bullosa disrupt
the binding interface.
The plakin domain is built up of nine spectrin repeats (SR1 to SR9) and a SH3 domain embedded in
the SR5. Using X-ray crystallography we have solved the structures of the regions SR1-SR2, SR3-SR4,
SR4-SR5-SH3, SR5-SR6, SR7-SR8, and SR7-SR9 (Sonnenberg et al J Mol Biol 2009;
Ortega et al J Biol Chem 2011;
Ortega et al J Biol Chem 2016).
We have used SAXS to elucidate the 3D structure of the SR3-SR9 region; which forms an extended
and slightly bent rod-like structure that spans ~340 Å.
Collectively our structures cover the repeats of the plakin domain and reveal significant structural variation along
these region.
Our contributions to the study of the structure of plectin are summarized in the following figure.
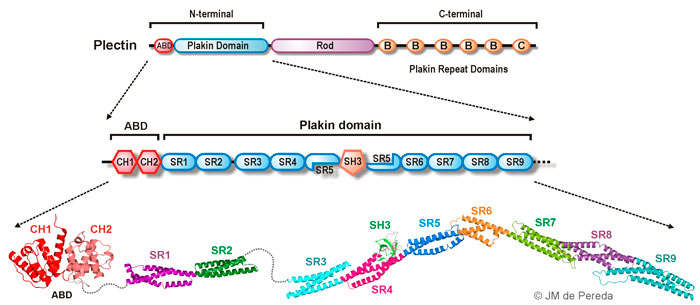
Structure of Plectin:. overall organization of the three major regions of plectin (top). Schematic representation of the sub-domains that compose the N-terminal region (middle). Ribbon diagrams of the 3D structures of the N-terminal region of plectin solved by our group, which includes the ABD and the complete plakin domain (SR1 to SR9).
